Research

Evolutionary biology of bacteria
Evolutionary genomics of bacteria
The increasing availability of bacterial genomes has opened the possibility for addressing fundamental questions of the evolutionary biology of bacteria: what does the genome wide variation can tell us about the demographic history of bacterial species? how has the gene composition of the genomes changed over time? how the acquisition of genes (outside of the common drug resistance genes) via lateral gene transfer has contributed to the adaptation of bacterial species to new niches?
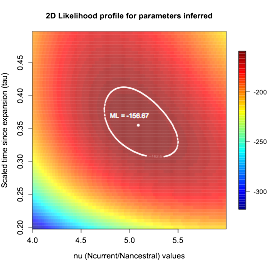
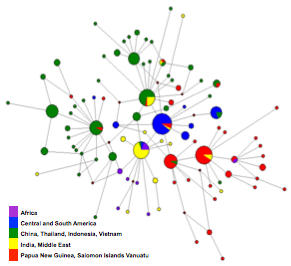
I have also performed demographic analyses with a more limited scope, aimed to understand the demographic history of Helicobacter pylori using a set of published genomes. How good are methods based on the site frequency spectrum at inferring the demographic history of bacteria that recombines frequently? I am in the process of writing a manuscript to present the results of this work.
I also maintain an active collaboration with my colleague Caitlin Pepperell at the University of Wisconsin-Madisson. Caitlin is interested in the evolution of pathogenic bacteria, specifically in Mycobacterium tuberculosis, and I have collaborated with her on the analysis of TB genomes using novel approaches, while our mutual overlap at Stanford. We plan on continuing our collaboration beyond TB and try to understand the impact of horizontal gene transfer in bacterial populations.
Biology of recombination in bacteria
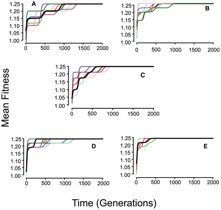
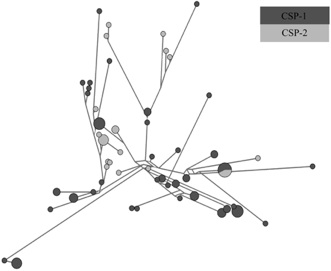
Additional work performed by Benjamin Evans, a former student of Danny Rozen has shown results that are consistent with our original assessment of the impact of the polymorphism on the population substructure of S. pneumoniae. We are preparing a manuscript presenting the results of this research.
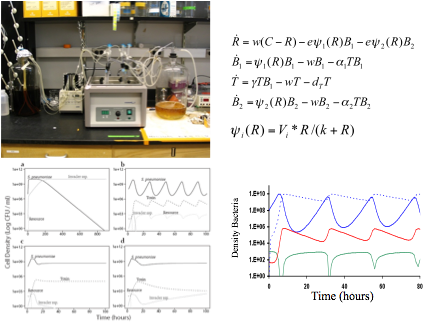
Ecology and evolution of bacteriocins
This work, at the intersection of experiments and mathematical modeling has also opened the door for an interesting hypothesis about the maintenance of self-killing agents with a set of testable hypotheses: if the toxin producing bacteria kills others more than it kill itself, then such a system could potentially be maintained. Nevertheless, as with the advantage of recombination, this mechanism engenders a cost to the organism and its action is frequency or density-dependent, making the question about its origin and maintenance very controversial. It seems that this mechanism could easily be maintained while common, but it would have problems at invading when rare. The question is not rhetorical, as for any strategy of biotic control of infections by symbiotic or carrier organisms, or intervention and control of contaminated cultures (i.e in dairy industry or cocoa fermentation), it would be ideal to count on bacteriocins that kill the organisms that produce them as well: once the contaminants have been accounted for, there is no need for the organism that produces the toxin.
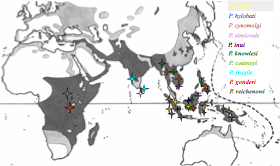
Population genetics and Phylogenetics of Plasmodium
Our interest in understanding the demographic history of Plasmodium vivax is not only an academic exercise, but also a necessary step towards understanding the ecological scenario in which selection has acted and how adaptation to the human host has historically occurred, and how drug resistance rises and spread in the population. Also, because recombination is tight to transmission, understanding the dynamics of seasonal epidemics and the overall historical changes will eventually help us make predictions on the speed at which adaptive evolution can occur in this species.
most of the analysis we have done so far have used a single locus (mitochondrial genomes) and I am planning on moving to full genomes analysis of Plasmodium vivax to better understand the demography and signatures of selection in this organism.
Evolutionary Genomics of Theobroma cacao
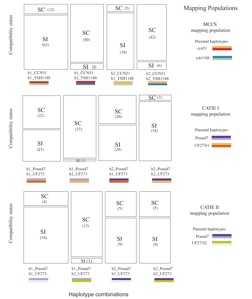
Previous work of our collaborator Juan Carlos Motamayor has shown that there are at least 10 main groups of genetically differentiated populations of cacao. This work has been based on the analysis of microsatellite markers. We have finished generating full genome sequence of accessions from 150 plants from all the groups identified with STRs to investigate in more detail the evolutionary history of cacao and identify the genetic basis of phenotypic traits. Only a small group of cacao plants have been used for breeding programs worldwide, thus identifying genomic resources among natural genotypes of cacao that could be of use in breeding programs is uttermost importance. Also, we would like to develop cacao as a model system for how to map complex traits in long-lived tropical trees where generating multi generational mapping populations is not feasible.
Cacao crops have an enormous economic impact, not only on the economy of chocolate producing countries, but also and most importantly on the economy of cocoa producing countries. Having a positive impact in society out of my research is as important to me as satisfying my intellectual curiosity and this project has a great potential of benefiting people in the long term. Cacao is an important crop, with an enormous economic impact for the producing countries (that are generally developing economies), and the principal motivation behind this research is the need to develop genomic resources to preserve cacao diversity and ensure a sustainable future for the crop. Also, we would like to develop cacao as a model system for how to map complex traits in long-lived tropical trees where generating multi generational mapping populations is not feasible.
maintained by O. Cornejo (last update 09/03/2012)